[ad_1]
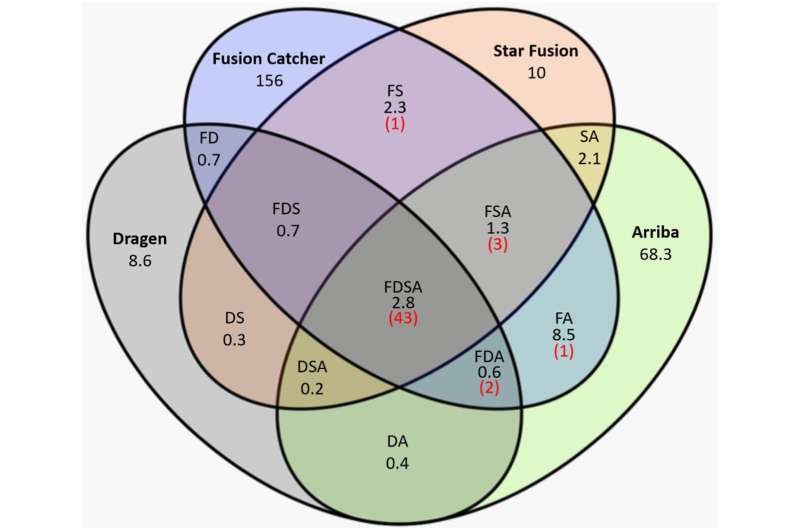
Venn diagram of imply variety of all fusions (in black) referred to as by the 4 callers, and the imply variety of reportable constructive fusion calls (in pink). A, Arriba; D, Dragen; F, FusionCatcher; S, STAR-Fusion. Credit score: The Journal of Molecular Diagnostics (2023). DOI: 10.1016/j.jmoldx.2023.11.003
Identification of particular gene fusions is essential for the profitable focused remedy of pediatric most cancers sufferers. Researchers at Youngsters’s Hospital Los Angeles have developed a novel assay that routinely integrates the information from a number of fusion identification instruments (callers) and effectively and precisely identifies clinically related gene fusions in pediatric tumors.
Their outcomes are reported in The Journal of Molecular Diagnostics.
Gene fusions are an vital class of driver alterations in most cancers that happen when components of two totally different genes be part of. Quite a few gene fusions have been recognized throughout several types of most cancers, particularly in pediatric malignant cancers. For instance, the PAX3-FOXO1 fusion gene is present in alveolar rhabdomyosarcoma however not in embryonal rhabdomyosarcoma.
Focused testing for gene fusions of curiosity on the DNA or RNA stage is presently in clinical use because it has diagnostic, prognostic, and therapeutic significance.
Lead investigator Jonathan Buckley, MD, Middle for Personalised Medication, Division of Pathology and Laboratory Medication, Youngsters’s Hospital Los Angeles; and Keck Faculty of Medication of USC, explains, “Correct identification of gene fusions in pediatric tumors is essential for establishing a prognosis and figuring out optimum remedy.”
“Sadly, the main software program packages for calling fusions, primarily based on genomic evaluation of tumor tissue, yield many false positives and/or fusions of no medical significance, they usually differ considerably within the units of fusions referred to as. Subsequently, there’s a compelling want for a software to help the medical molecular geneticist to filter and prioritize the fusion calls to maximise effectivity and accuracy.”
Dr. Buckley and his co-investigators developed, examined, and validated a brand new exome seize–primarily based RNA-sequencing (RNAseq) assay. They used the Twist Bioscience Complete Exome seize equipment with RNA from contemporary, frozen, or formalin-fixed samples from sufferers with pediatric hematologic malignancies and strong tumors and a gene fusion detection assay that employs 4 fusion callers (Arriba, FusionCatcher, STAR-Fusion, and Dragen).
The brand new bioinformatics platform detects fusions, prioritizes them, and customized curates downstream processes for consensus fusion calling with excessive accuracy and effectivity.
Whereas the 4 fusion-caller software program packages are in extensive use, their outcomes usually embody a lot of false-positive calls and/or identification of fusions of no medical worth. This presents a major problem in medical reporting, each by way of effectivity and accuracy.
The investigators had been initially stunned to notice the variations among the many units of fusions recognized by the 4 callers given an identical RNAseq information processes, related approaches, and the identical finish purpose.
Dr. Buckley famous, “We had been pleasantly stunned to find out that integration of information from the 4 callers, and utility of advert hoc rating strategies primarily based on each the genomic information and references sources (associated to the reported fusions and their element genes), was extremely efficient in highlighting the important thing medical fusion(s).”
The software program proved extremely efficient in pinpointing recognized clinically related fusions, rating them first in 47 of fifty (94%) samples. To spotlight the usefulness of the brand new assay and to indicate the significance of a genome-wide and nontargeted technique for fusion detection in pediatric most cancers, the researchers current findings from three diagnostically difficult circumstances.
The present OncoKids DNA and RNA-based next-generation sequencing (NGS) assay used on the middle didn’t establish driver gene fusions in these sufferers. Nevertheless, the brand new technique discovered disease-related gene fusions that had been then confirmed by different established sequencing checks.
Dr. Buckley famous, “The complete spectrum of gene fusions that operate as drivers of tumorigenesis in pediatric tumors continues to be unknown. One gene can fuse with one and even a whole bunch of associate genes with the identical web biologic impact. A genome-wide strategy is due to this fact crucial when the anticipated or widespread gene fusions usually are not recognized by routine methodologies.”
Senior creator Jaclyn A. Biegel, Ph.D., Youngsters’s Hospital Los Angeles and Keck Faculty of Medication of USC, concluded, “Our research exhibit that the mixture of an exome seize–primarily based strategy for RNAseq and a bioinformatics platform to streamline the evaluation is each sturdy and environment friendly to precisely establish pathogenic gene fusions in bone marrow samples in addition to contemporary, frozen, and formalin-fixed tissue specimens from pediatric most cancers sufferers.”
“Though we validated this for medical use for childhood tumors, these approaches can simply be prolonged to grownup sufferers with hematologic malignancies and strong tumors.”
Extra info:
Jonathan Buckley et al, An Exome Seize-Primarily based RNA-Sequencing Assay for Genome-Huge Identification and Prioritization of Clinically Vital Fusions in Pediatric Tumors, The Journal of Molecular Diagnostics (2023). DOI: 10.1016/j.jmoldx.2023.11.003
Quotation:
New assay identifies clinically related gene fusions in pediatric tumors extra precisely and effectively (2024, February 13)
retrieved 13 February 2024
from https://medicalxpress.com/information/2024-02-assay-clinically-relevant-gene-fusions.html
This doc is topic to copyright. Aside from any truthful dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is offered for info functions solely.
[ad_2]
Source link




Discussion about this post