[ad_1]
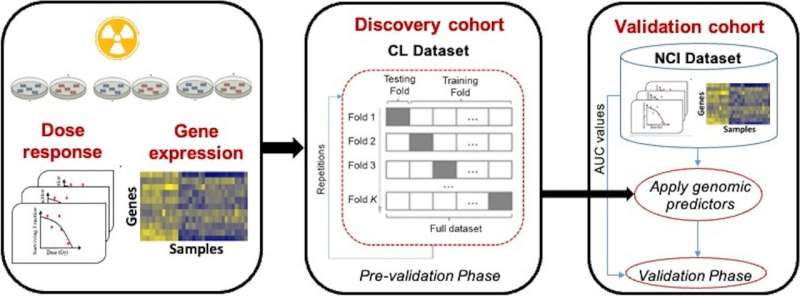
Evaluation pipeline. Utilizing the cross-validation framework within the discovery cohort, CL dataset, we carried out the pre-validation of genomic predictors for radiation sensitivity. Gene expression signature was developed utilizing the total coaching set, which was then evaluated on a completely impartial exterior cohort, NCI cohort. Credit score: BMC Most cancers (2024). DOI: 10.1186/s12885-023-11634-3
Because the effectiveness of radiotherapy varies vastly between tumors, discovering gene signatures to foretell the radiation response may higher information clinicians to personalize therapy plans.
Professor Venkata Manem, affiliated with the School of Medication at Université Laval and the Middle de recherche CHU de Québec—Université Laval, has made a promising step to advance pre-clinical analysis within the area of precision radiation oncology. The examine is revealed within the journal BMC Cancer.
Presently, medical doctors use a “one-size-fits-all” paradigm for radiotherapy, with a given dose and frequency of therapy, whatever the tumor’s genomic options.
“Some cancers can be extra delicate or extra proof against several types of radiation routine. By figuring out sufferers who can profit from decrease doses, we may cut back the therapy’s toxicity. Whereas on the identical time, we may regulate the dose for extra resistant tumors, or mix it with different therapies,” explains Manem, previously an assistant professor on the Université du Québec à Trois-Rivières.
For now, the developed radiosensitivity markers could be utilized throughout all cancers typically, however the group is hoping to construct biomarkers which can be tissue particular. “With the provision of tissue-specific knowledge, we may finally have signatures for several types of cancers reminiscent of breast, prostate and lung cancers,” says Professor Venkata Manem.
“All tumors are completely different, even when they’re categorized in the identical group, on the identical stage, and with the identical anatomical options. They differ in a number of facets, reminiscent of mutations current, microenvironment, and immune part. All these elements can have an effect on the radiation response,” provides Alona Kolnohuz, first writer of the examine.
Utilizing cell line knowledge coupled with bioinformatics and machine learning-based approaches, the analysis group has constructed a molecular predictor of radiation response that might be examined in a pre-clinical setting, earlier than being carried out in clinical studies.
“Most research within the area use the variety of cells that survive at a given radiation dose, say at 2Gy of radiation dose, which is equal to a single level to conclude. Our method makes use of the realm below the radiation dose-response curve (AUC) as a substitute. Based mostly on our findings, we concluded that AUC needs to be thought-about as a pre-clinical radioresponse indicator in future research because it captures a wider vary of organic processes,” explains Professor Manem.
The following stage of his analysis includes validating the developed molecular signature in affected person knowledge and constructing a scientific assay utilizing interpretable machine studying based mostly strategies together with figuring out radiosensitizing compounds that may enhance the therapeutic efficacy of radiation.
“With the rising OMICS and AI-driven applied sciences, time is now ripe for precision medication to take an enormous leap away from the standard ‘one-size-fits-all’ framework,” says Venkata Manem. We envision that the developed gene signature of radiation sensitivity has the large potential to assist decision-making, personalize therapies, and enhance outcomes.
Extra data:
Alona Kolnohuz et al, Gene expression signature predicts radiation sensitivity in cell strains utilizing the integral of dose–response curve, BMC Most cancers (2024). DOI: 10.1186/s12885-023-11634-3
Offered by
Laval University
Quotation:
Figuring out genomics markers to foretell radiation sensitivity (2024, February 2)
retrieved 4 February 2024
from https://medicalxpress.com/information/2024-02-genomics-markers-sensitivity.html
This doc is topic to copyright. Other than any honest dealing for the aim of personal examine or analysis, no
half could also be reproduced with out the written permission. The content material is supplied for data functions solely.
[ad_2]
Source link




Discussion about this post