[ad_1]
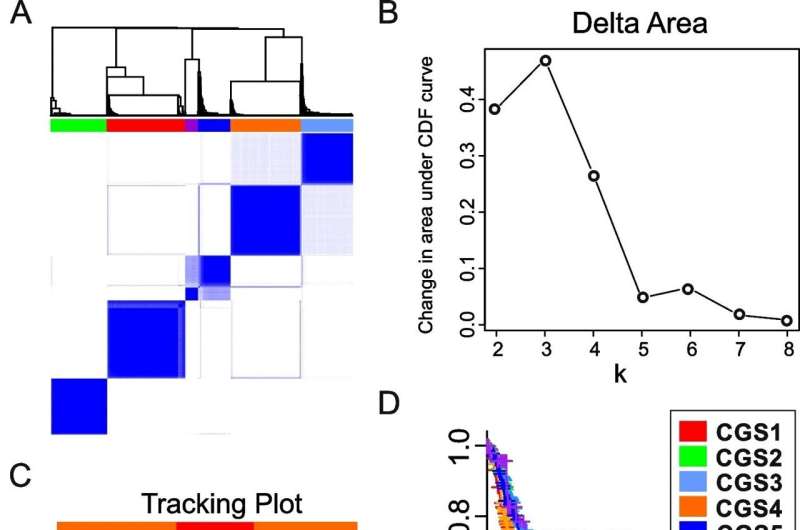
Consensus subtypes of gastric adenocarcinoma found by cluster-of-cluster task. A Hierarchical dendrogram of supercluster subtypes recognized by cluster-of-cluster task (COCA). GAC tumors have been grouped in line with shared options of independently found molecular subtypes. Tumors from 6 GAC cohorts (Korea, Samsung, TCGA, Shanghai, Shanxi, and Singapore1, n = 1427) have been stratified by 8 genomic classification strategies, and subtype info was used for clustering with ConsensusClusterPlus (v.3.12). Columns and rows characterize GAC tumors within the discovery/coaching set. B Delta space plot. The plot reveals the relative change in space beneath the cumulative distribution perform (CDF) curve evaluating okay and okay − 1. C Monitoring plot. The plot reveals the cluster task of tumors (columns) for every okay (rows) by shade. This plot offers a view of merchandise cluster membership throughout completely different values of okay. D Medical significance of 6 consensus subtypes of GAC in discovery/coaching cohorts (n = 1185). Sufferers within the Korea, TCGA, Shanxi, and Samsung cohorts have been included within the evaluation; scientific information from the Shanghai and Singapore1 cohorts weren’t obtainable. OS, total survival. P-value signifies the importance from log-rank check. Credit score: Molecular Most cancers (2023). DOI: 10.1186/s12943-023-01796-w
A analysis crew has introduced a brand new genetic classification system for gastric most cancers by a multicenter research with the College of Texas MD Anderson Most cancers Heart. This analysis might lay the inspiration for personalised gastric most cancers remedy.
5 establishments together with the MD Anderson Most cancers Heart, Korea College Faculty of Drugs, CHA College Medical Heart, Kyung Hee College Faculty of Drugs, Yonsei College Faculty of Drugs, and Sungkyunkwan College Faculty of Drugs; participated on this multicenter research.
The analysis, published within the journal Molecular Most cancerswas led by Professor Ju‑Seog Lee of MD Anderson Most cancers Heart and Professor Sang Cheul Oh of Division of Oncology/Hematology participated. Professor Sang‑Hee Kang of Division of Surgical procedure and Professor Solar Younger Yim of Division of Gastroenterology and Hepatology participated led because the co-lead authors.
Beforehand, the MD Anderson- Korea College analysis crew additionally printed the genetic classification system for liver cancer in Hepatology.
Gastric cancer tends to have genetic and scientific variety. The analysis crew analyzed the gene classification system for 8 gastric cancers that was printed prior to now and derived the results of 6 Consensus Genomic Subtypes (CGSs). This method categorized gastric cancers from CGS1 to CGS6 relying on completely different gene expression patterns.
Every subtype had completely different traits and CGS1 confirmed the worst prognostic options. It had very excessive stem cell properties and low genetic modification. Nonetheless, the evaluation confirmed that CGS1 responded properly to immunotherapy and coverings concentrating on IGF1R could possibly be efficient. CGS2 was enriched in typical epithelial cell gene expression. CGS3 and CGS4 confirmed excessive cloning quantity variation and had low responses to immunotherapy.
Nonetheless, CGS3 had a attribute of HER2 gene activation and CGS4 had a attribute of SALL4 gene activation. The crew analyzed that remedies concentrating on these options can be efficient. CGS5 had excessive mutation burden, which is a attribute of microsatellite instability in tumors, and confirmed reasonable response to immunotherapy. CGS6 was largely constructive for infectious mononucleosis (Epstein Barr) virus and had very excessive methylation ranges. It confirmed a excessive response to immunotherapy.
The analysis crew not solely categorized genetics of gastric cancerbut in addition estimated the potential response charges of ordinary and experimental remedies (chemoradiotherapy, immunotherapy, and many others.) for every subtype by systematic evaluation of genome and proteome information. Because of this, the CGS3 subtype confirmed particularly nice advantages in anticancer radiotherapy on account of its iron-dependent cell loss of life on account of excessive degree lipid peroxidation. Analysis instructed potential remedy topics for every subtype.
Professor Solar Younger Yim of the Division of Gastroenterology and Hepatology, one of many lead authors of this research, stated, “Though the mortality charge of gastric most cancers is reducing as a result of implementation of recent therapies, it nonetheless is likely one of the main causes of loss of life for cancer patients. We imagine that this analysis goes to put the inspiration for personalised gastric most cancers remedies.”
Extra info:
Yun Seong Jeong et al, Clinically conserved genomic subtypes of gastric adenocarcinoma, Molecular Most cancers (2023). DOI: 10.1186/s12943-023-01796-w
Supplied by
Korea College Faculty of Drugs
Quotation:
A brand new gene classification system for gastric most cancers (2023, November 6)
retrieved 7 November 2023
from https://medicalxpress.com/information/2023-11-gene-classification-gastric-cancer.html
This doc is topic to copyright. Other than any truthful dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is offered for info functions solely.
[ad_2]
Source link




Discussion about this post